The Bioinformatics and Analytics Core distributes data for the Genomics Technology Core and our own clients through a data distribution system. If you have received an email from our team that your data is available, you can use the information below to help you access your data. If you have any problems accessing your data, please contact us at bioinformatics@missouri.edu.
Account Creation
MU Clients
For clients from the MU System you will use your SSO (Single Sign On) login username and password to access your data. These are the same credentials that you use to access your other MU services.
External Clients
For external clients, you should have received an email that contains your username. If you are a first time user, you will first need to visit the account setup site to create your password. For account creation, please make sure to use the email address you requested the data be distributed to when completing your project authorization with the GTC. All your data will be made available through this same account as long as you continue using this email address. If you can’t remember that address, please contact mugenomicscore@missouri.edu for help additional help.
Web Access
Once you have your account credentials, you can visit https://download.bioinformatics.missouri.edu in order to browse and download your data through a web browser. This feature is made available for convenience, however due to the size of sequencing data, it is highly advised you download your data using one of the methods described below (SFTP or Rsync).
SFTP
Secure File Transfer Protocol (SFTP) is a method to transfer that uses SSH encryption to transfer files between systems securely. SFTP supports the full feature set of FTP, as well as additional features. Users can use SFTP via command line or several different software packages with a graphical interface to transfer files.
MU System Clients
The University of Missouri-Columbia campus recommends using the SecureFX SFTP client. This client is made available on Windows systems via Software Center to anyone on campus by the Division of IT. Please contact doit.missouri.edu or your local IT pro for help installing a supported SFTP client. Installation of SFTP clients not found in Software Center will require administrator access on your system.
Using SecureFX
Connect
1. Start SecureFX by clicking on the icon on your desktop, under the start menu or on your dock.
2. The Connect window should display. If not, go to the File menu and click Connect.
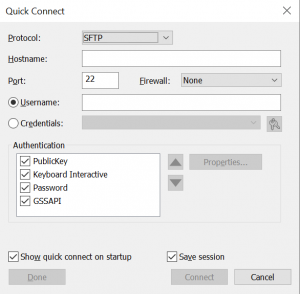
3. The first time you connect to a new session, the New Host Key window may appear. Click Accept & Save.
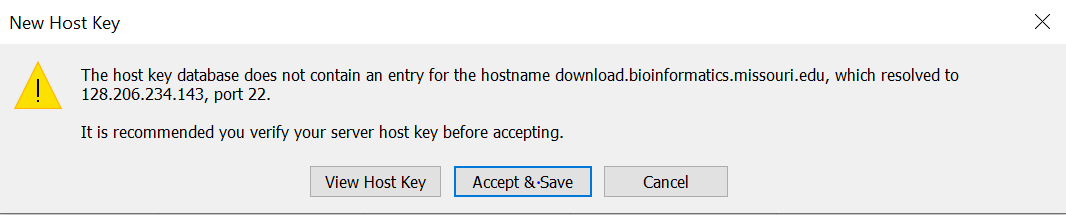
4. Enter your username in the username field and then click OK.
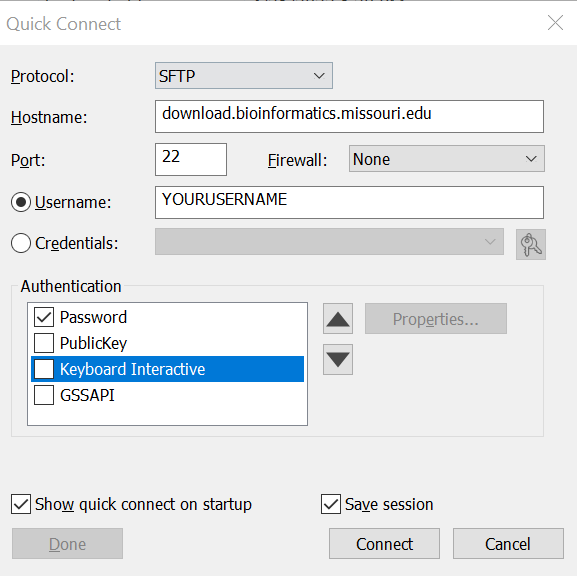
5. Enter your account password in the password field and click OK. Do not check the box to save your password.
6. You should now be connected.
Downloading
- You will see your data on the Bioinformatics and Analytics Core server on the right-hand tab. Your local computer is on the left hand tab.
- This lets you easily drag and drop files between your desktop and the remote server. You can now drag and drop your data from our servers to your local computer to download them.
- Please note: sequencing data is often quite large, and can take a long time to download. Make sure you have a stable internet connection and be prepared to allow your computer to transfer data for several hours or days in many cases. Also, please make sure you have enough storage space on your local system to save your data. It is not unusual for sequencing data to be in the hundreds of gigabytes to several terabyte size range. Many laptop and desktop computers will not have enough storage space to save your data unless you have an external storage drive attached.
Disconnect
- To disconnect from a session, close the session window. Or, go to the File menu and click Disconnect.
- To exit SecureFX, go to the File menu and click Exit.
Other SFTP Clients
For external users, or those who have access to or prefer to use a different SFTP client you’ll need to follow the connection guides for your particular client. The settings provided in your data distribution email should work with most clients, but you may have to look in different locations within the software to change those specific settings.
A few clients we have tested successfully include:
- Filezilla (Free and Open Source – Windows, Mac, and Linux)
- CyberDuck (Free / Paid – Windows and Mac)
- Transmit (Paid – Mac)
- WinSCP (Free – Windows)
RSYNC
Rsync or “Remote Sync”, is a free command-line tool that lets you transfer files and directories to local and remote destinations. Rsync is used for mirroring, performing backups, or migrating data to other servers. It is fast and efficient with the ability to only copy the differences between the source and and destination files. Rsync is natively installed on Mac and Linux operating systems. For Windows you will need to install WSL in order to use Rsync.
Transferring your data to the MU HPC Cluster System (Lewis)
For MU researchers and affiliates, you can use Rsync to transfer your data to the Lewis cluster system over the DTN (data transfer node). If you are new to using the Lewis cluster, please reach out to Research Support Solutions for help in getting your account set up. If you are transferring data to Lewis, Rsync is the BAC / GTC recommended way to copy your data to Lewis. In your data distribution email, you should have received a pre-generated rsync command that you can use to download your data for your convenience.
